Representation of a microarray dataset that is used by Seek. More...
#include <seekdataset.h>
Public Types | |
| enum | DistanceMeasure { CORRELATION = 0, Z_SCORE = CORRELATION + 1 } |
| Distance measure (see main section for descriptions) More... | |
Public Member Functions | |
| CSeekDataset () | |
| Constructor. | |
| ~CSeekDataset () | |
| Destructor. | |
| bool | ReadDatasetAverageStdev (const string &) |
Read the *.sinfo file. | |
| bool | ReadGeneAverage (const string &) |
Read the gene average correlation file *.gavg. | |
| bool | ReadGeneVariance (const string &) |
Read the gene variance file *.gvar. | |
| bool | ReadGenePresence (const string &) |
Read the gene presence file *.gpres. | |
| bool | InitializeGeneMap () |
| Initialize the genome presence map. | |
| bool | InitializeQuery (const vector< utype > &) |
| Initialize the query presence map. | |
| bool | InitializeQueryBlock (const vector< utype > &) |
| Initialize a presence map for a block of queries. | |
| bool | DeleteQuery () |
| Delete the query. | |
| bool | DeleteQueryBlock () |
| Delete query block. | |
| bool | InitializeDataMatrix (utype **, const vector< float > &, const utype &, const utype &, const bool=true, const bool=false, const bool=false, const enum DistanceMeasure=Z_SCORE, const float cutoff=-1.0 *CMeta::GetNaN(), const bool=false, gsl_rng *rand=NULL) |
| Initialize the gene-gene correlation matrix. | |
| bool | Copy (CSeekDataset *) |
| Copy constructor. | |
| utype ** | GetDataMatrix () |
| Get the gene-gene correlation matrix. | |
| unsigned char ** | GetMatrix () |
| Get the gene-gene correlation matrix. | |
| CSeekIntIntMap * | GetGeneMap () |
| Get the genome presence map. | |
| CSeekIntIntMap * | GetDBMap () |
| Get the query-block presence map. | |
| CSeekIntIntMap * | GetQueryMap () |
| Get the query presence map. | |
| const vector< utype > & | GetQuery () const |
| Get the query genes. | |
| const vector< utype > & | GetQueryIndex () const |
| Get the query gene indices. | |
| float | GetGeneVariance (const utype &) const |
| Get the gene expression variance vector. | |
| float | GetGeneAverage (const utype &) const |
| Get the gene average correlation vector. | |
| float | GetDatasetAverage () const |
| Get the mean of the global gene-gene Pearson distribution. | |
| float | GetDatasetStdev () const |
| Get the standard deviation of the global gene-gene Pearson distribution. | |
| utype | GetNumGenes () const |
| Get the genome size. | |
| bool | InitializeCVWeight (const utype &) |
| Initialize the weight of the dataset. | |
| bool | SetCVWeight (const utype &, const float &) |
| Set the score for a particular cross-validation. | |
| float | GetCVWeight (const utype &) |
| Get the score for a particular cross-validation. | |
| const vector< float > & | GetCVWeight () const |
| Get all the cross-validation scores. | |
| float | GetDatasetSumWeight () |
| Get the dataset weight. | |
| void | SetPlatform (CSeekPlatform &) |
| Set the platform. | |
| CSeekPlatform & | GetPlatform () const |
| Get the platform. | |
Detailed Description
Representation of a microarray dataset that is used by Seek.
A CSeekDataset encapsulates the following information about the dataset:
- The gene-gene correlation matrix
- Each gene's expression variance
- Each gene's average correlation
- The genes in the query that are present in the dataset
- The genes in the genome that are present in the dataset
- The platform which the dataset belongs to
- The weight of the dataset that is assigned by the search algorithm
This dataset structure is designed to be used by Seek.
- Remarks:
- The word correlation refers to the standardized z-scores of Pearson correlations, which is derived from a 2-step process:
where![\[f(x,y)=\frac{1}{2}ln\frac{1+p(x,y)}{1-p(x,y)}\]](form_2.png)
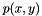 is the Pearson correlation,
is the Pearson correlation, 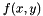 is the Fisher's transformed score.
is the Fisher's transformed score.
where![\[z(x,y)=\frac{f(x,y) - \bar{f}}{\sigma_{f}}\]](form_9.png)
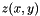 is the z-score,
is the z-score, 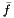 is the mean, and
is the mean, and 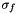 is the standard deviation.
is the standard deviation.
From here on, correlation always refers to the above z-score definition.
Definition at line 133 of file seekdataset.h.
Member Enumeration Documentation
Distance measure (see main section for descriptions)
Definition at line 140 of file seekdataset.h.
Member Function Documentation
| bool Sleipnir::CSeekDataset::Copy | ( | CSeekDataset * | src | ) |
| bool Sleipnir::CSeekDataset::DeleteQuery | ( | ) |
Delete the query.
Resets all query-related data, such as dataset weight, query presence map, etc.
Definition at line 240 of file seekdataset.cpp.
Referenced by DeleteQueryBlock(), InitializeQuery(), and ~CSeekDataset().
Delete query block.
Resets all query-block related data.
Definition at line 255 of file seekdataset.cpp.
References DeleteQuery(), and Sleipnir::CSeekTools::Free2DArray().
Referenced by InitializeQueryBlock(), and ~CSeekDataset().
| float Sleipnir::CSeekDataset::GetCVWeight | ( | const utype & | i | ) |
Get the score for a particular cross-validation.
- Parameters:
-
i The index
Definition at line 572 of file seekdataset.cpp.
| const vector< float > & Sleipnir::CSeekDataset::GetCVWeight | ( | ) | const |
Get all the cross-validation scores.
- Returns:
- A vector of cross-validation scores
Definition at line 576 of file seekdataset.cpp.
| utype ** Sleipnir::CSeekDataset::GetDataMatrix | ( | ) |
Get the gene-gene correlation matrix.
- Returns:
- A two-dimensional array of type
utype. Note that the correlation has been scaled to a integer range from 0 to 640. See CSeekDataset::InitializeDataMatrix.
Definition at line 277 of file seekdataset.cpp.
Referenced by Sleipnir::CSeekWeighter::LinearCombine().
| float Sleipnir::CSeekDataset::GetDatasetAverage | ( | ) | const |
Get the mean of the global gene-gene Pearson distribution.
- Returns:
- The mean Pearson for the dataset
Definition at line 540 of file seekdataset.cpp.
| float Sleipnir::CSeekDataset::GetDatasetStdev | ( | ) | const |
Get the standard deviation of the global gene-gene Pearson distribution.
- Returns:
- The standard deviation of the Pearson distribution
Definition at line 544 of file seekdataset.cpp.
Get the query-block presence map.
- Returns:
- The query-block presence map
Definition at line 536 of file seekdataset.cpp.
| float Sleipnir::CSeekDataset::GetGeneAverage | ( | const utype & | i | ) | const |
Get the gene average correlation vector.
- Returns:
- The average correlation vector
Definition at line 552 of file seekdataset.cpp.
Referenced by InitializeDataMatrix().
Get the genome presence map.
- Returns:
- The genome presence map
Definition at line 528 of file seekdataset.cpp.
Referenced by Sleipnir::CSeekWeighter::CVWeighting(), Sleipnir::CSeekWeighter::LinearCombine(), and Sleipnir::CSeekWeighter::OneGeneWeighting().
| float Sleipnir::CSeekDataset::GetGeneVariance | ( | const utype & | i | ) | const |
Get the gene expression variance vector.
- Returns:
- The variance vector
Definition at line 548 of file seekdataset.cpp.
| unsigned char ** Sleipnir::CSeekDataset::GetMatrix | ( | ) |
Get the gene-gene correlation matrix.
- Returns:
- A two-dimensional array of type
unsignedchar**.
Definition at line 524 of file seekdataset.cpp.
| utype Sleipnir::CSeekDataset::GetNumGenes | ( | ) | const |
Get the genome size.
- Returns:
- The genome size
Definition at line 556 of file seekdataset.cpp.
Referenced by Sleipnir::CSeekWeighter::CVWeighting(), Sleipnir::CSeekWeighter::LinearCombine(), and Sleipnir::CSeekWeighter::OneGeneWeighting().
| CSeekPlatform & Sleipnir::CSeekDataset::GetPlatform | ( | ) | const |
Get the platform.
- Returns:
- The platform of this dataset
Definition at line 599 of file seekdataset.cpp.
| const vector< utype > & Sleipnir::CSeekDataset::GetQuery | ( | ) | const |
| const vector< utype > & Sleipnir::CSeekDataset::GetQueryIndex | ( | ) | const |
Get the query gene indices.
- Returns:
- A vector of query gene indices
Definition at line 273 of file seekdataset.cpp.
Get the query presence map.
- Returns:
- The query presence map
Definition at line 532 of file seekdataset.cpp.
Referenced by Sleipnir::CSeekWeighter::CVWeighting(), Sleipnir::CSeekWeighter::LinearCombine(), and Sleipnir::CSeekWeighter::OneGeneWeighting().
| bool Sleipnir::CSeekDataset::InitializeCVWeight | ( | const utype & | i | ) |
Initialize the weight of the dataset.
- Parameters:
-
i The number of cross-validations
Initializes the total dataset weight, and the score of the individual cross-validation (CV) runs.
Definition at line 560 of file seekdataset.cpp.
Referenced by Sleipnir::CSeekWeighter::CVWeighting(), and Sleipnir::CSeekWeighter::OneGeneWeighting().
| bool Sleipnir::CSeekDataset::InitializeDataMatrix | ( | utype ** | , |
| const vector< float > & | , | ||
| const utype & | , | ||
| const utype & | , | ||
| const bool | = true, |
||
| const bool | = false, |
||
| const bool | = false, |
||
| const enum | DistanceMeasure = Z_SCORE, |
||
| const float | cutoff = -1.0*CMeta::GetNaN(), |
||
| const bool | = false, |
||
| gsl_rng * | rand = NULL |
||
| ) |
Initialize the gene-gene correlation matrix.
- Parameters:
-
rD A two-dimensional array storing the discretized gene-gene correlations quant The discretization function iRows The number of rows for the correlation matrix iColumns The number of columns for the correlation matrix bSubtractAvg If true, subtract the correlation by the dataset average bNormPlatform If true, subtract the correlation by the platform average and divide by standard deviation logit If true, apply the logit transform on correlations dist_measure Distance measure: z-score or correlations cutoff Apply a hard cutoff on correlations bRandom If true, shuffle the correlation vector rand The random generator for the shuffling operation above
- Remarks:
- The discretized correlation in the matrix
rDis bounded by 0 to 255 (the limit ofunsignedchar). The parameterquantspecifies how a correlation is discretized. For example, if thequanthas 5 bins:Then if a correlation is 2.5, the discretized value would be 2.[0, 1, 2, 3, 4]
Definition at line 281 of file seekdataset.cpp.
References CORRELATION, Sleipnir::CSeekTools::Free2DArray(), Sleipnir::CSeekIntIntMap::GetAllReverse(), GetGeneAverage(), Sleipnir::CSeekIntIntMap::GetNumSet(), Sleipnir::CSeekPlatform::GetPlatformAvg(), Sleipnir::CSeekPlatform::GetPlatformStdev(), Sleipnir::CSeekIntIntMap::GetReverse(), Sleipnir::CSeekTools::Init2DArray(), and Sleipnir::CSeekTools::InitVector().
Initialize the genome presence map.
Indicates which genes of the genome are present in the dataset.
Definition at line 132 of file seekdataset.cpp.
References Sleipnir::CSeekIntIntMap::Add(), and Sleipnir::CMeta::IsNaN().
| bool Sleipnir::CSeekDataset::InitializeQuery | ( | const vector< utype > & | query | ) |
Initialize the query presence map.
- Parameters:
-
query The query genes
Indicates which query genes are present in the dataset.
Definition at line 190 of file seekdataset.cpp.
References Sleipnir::CSeekIntIntMap::Add(), DeleteQuery(), Sleipnir::CSeekIntIntMap::GetForward(), and Sleipnir::CSeekTools::IsNaN().
| bool Sleipnir::CSeekDataset::InitializeQueryBlock | ( | const vector< utype > & | queryBlock | ) |
Initialize a presence map for a block of queries.
- Parameters:
-
queryBlock A vector of queries
Flattens all the queries into one vector that contains only the unique query genes, then constructs a presence map based on this vector.
Definition at line 154 of file seekdataset.cpp.
References Sleipnir::CSeekIntIntMap::Add(), DeleteQueryBlock(), Sleipnir::CSeekIntIntMap::GetForward(), Sleipnir::CSeekIntIntMap::GetNumSet(), Sleipnir::CSeekTools::Init2DArray(), and Sleipnir::CSeekTools::IsNaN().
| bool Sleipnir::CSeekDataset::ReadDatasetAverageStdev | ( | const string & | strFileName | ) |
Read the *.sinfo file.
- Parameters:
-
strFileName The file name
The *.sinfo file contains the mean and the standard deviation of the global gene-gene Pearson distribution for this dataset.
Definition at line 111 of file seekdataset.cpp.
References Sleipnir::CSeekTools::ReadArray().
| bool Sleipnir::CSeekDataset::ReadGeneAverage | ( | const string & | strFileName | ) |
Read the gene average correlation file *.gavg.
- Parameters:
-
strFileName The file name
The *.gavg is an array that stores the average correlation of each gene.
Definition at line 119 of file seekdataset.cpp.
References Sleipnir::CSeekTools::ReadArray().
| bool Sleipnir::CSeekDataset::ReadGenePresence | ( | const string & | strFileName | ) |
Read the gene presence file *.gpres.
- Parameters:
-
strFileName The file name
The *.gpres is a 2-value array that contains the presence (1), absence (0) status of genes.
Definition at line 123 of file seekdataset.cpp.
References Sleipnir::CSeekTools::ReadArray().
| bool Sleipnir::CSeekDataset::ReadGeneVariance | ( | const string & | strFileName | ) |
Read the gene variance file *.gvar.
- Parameters:
-
strFileName The file name
The *.gvar file is an array that stores the expression variance of each gene.
Definition at line 127 of file seekdataset.cpp.
References Sleipnir::CSeekTools::ReadArray().
| bool Sleipnir::CSeekDataset::SetCVWeight | ( | const utype & | i, |
| const float & | f | ||
| ) |
Set the score for a particular cross-validation.
- Parameters:
-
i The index f The validation score
Definition at line 567 of file seekdataset.cpp.
Referenced by Sleipnir::CSeekWeighter::CVWeighting(), and Sleipnir::CSeekWeighter::OneGeneWeighting().
| void Sleipnir::CSeekDataset::SetPlatform | ( | CSeekPlatform & | cp | ) |
The documentation for this class was generated from the following files:
- src/seekdataset.h
- src/seekdataset.cpp
 1.7.6.1
1.7.6.1